Using pre-trained prot_bert to predict protein attributes
The pre-trained attention-based language models have proved themselves super powerful. In life science field, models pre-trained on protein sequences can also extract a considerable amount of hidden information. A short note on BERTology in this blog.
Sometimes, just laying a very simple MLP on top of bert’s embedding can give us quite satisfactory results. Below are my notes on using prot_bert on a binary classification task.
Pre-trained models can be found from Huggingface. Since I use TensorFlow I need to load the weights from Rostlab/prot_bert into a keras model. In the transformers package it only takes one extra argument.
Model Implementation
First create the base model class.
class ProtBertBase(keras.models.Model):
def __init__(self, finetune: bool = False, *args, **kwargs):
super(ProtBertBase, self).__init__(*args, **kwargs)
self.tokenizer = TFBertTokenizer.from_pretrained("Rostlab/prot_bert") # 1
self.base_model = TFBertModel.from_pretrained("Rostlab/prot_bert", from_pt=True) # 2
self.base_model.trainable = finetune # 3
def call(self, inputs):
encoded = self.tokenizer(inputs)
embedding = self.base_model(**encoded)
# 4
pool_emb = tf.reduce_mean(
tf.ragged.boolean_mask(
embedding.last_hidden_state,
mask=tf.cast(
encoded['attention_mask'],
dtype=bool,
)
),
axis=1
)
outputs = self.top_model(pool_emb)
return outputs
# 5
@classmethod
def from_top_model_weights(cls, path, *args, **kwargs):
model = cls(*args, **kwargs)
model.top_model.load_weights(path)
return model
Subclass the base model class to add the top_model:
class MlpOnProtBertBase(ProtBertBase): # 6
"prot_bert embedding + simple MLP"
def __init__(self, units, *args, **kwargs):
super(MlpOnProtBertBase, self).__init__(*args, **kwargs)
self.top_model = keras.models.Sequential([
keras.layers.Dense(units, activation='relu'),
keras.layers.Dense(1, activation='sigmoid')
])
Let’s break down the codes by each comment item.
TFBertTokenizerInstead of preprocessing the sequences and save the embedding for modelling, we can use theTFTokenizeras a keras layer and process the embedding on the fly as its docstring says:In-graph tokenizers, unlike other Hugging Face tokenizers, are actually Keras layers and are designed to be run when the model is called, rather than during preprocessing.
- Load weights from PyTorch. Since rostlab/prot_bert only has the PyTorch weights we need to specify
from_pt=True. - The model is trainable by default. If we just need the embedding, we don’t need to pass gradients to base model. Only when we want to fine tune the base model we need to make the base model trainable.
- The tokenizer outputs a dict with 3 keys: ‘input_ids’, ‘attention_mask’, ‘token_type_ids’. By my experiments, masked embedding performs better (also will be discussed below).
- If the base model is not changed we don’t have to save the full model. This classmethod is used to retrieve weigths from the saved top model.
- By subclassing the
ProtBertBase, we only need to define thetop_model.
pooler_output vs. last_hidden_state
The embedding has 2 attributes, pooler_output with shape (batch, embedding_dim) and last_hidden_state with shape (batch, seq_max_len, embedding_dim). Here the embedding_dim is 1024.
Initially I used the pooler_output but the result was terrible. The model could not learn anything. Each batch was either all True or all False. After several trials I changed to pool the last_hidden_state by the seq_max_len dimension and it turned out to have quite good results.
A similar discussion mentioned that there is no universal standard. Better experiment on both.
Mask on pooled embedding
After I switched to pool the last_hidden_state for embedding, I just pooled it along the 2nd dimension (seq_max_len):
pool_emb = tf.math.reduce_mean(embedding.last_hidden_state, axis=1)
Since the sequence lengths are different, it is natural to think about applying the mask:
pool_emb = tf.reduce_mean(
tf.ragged.boolean_mask(
embedding.last_hidden_state,
mask=tf.cast(
encoded['attention_mask'],
dtype=bool,
)
),
axis=1
)
The trick here is the tf.ragged.boolean_mask which will return a ragged tensor by removing elements masked. Here’s a simple example:
var = tf.Variable([
[[0.5, 0.5], [0.8, 0.8], [1.0, 1.0]],
[[0.2, 0.2], [0.4, 0.4], [1.0, 1.0]],
])
# Shape: (batch, seq_len, emb_dim)
var.shape
"""
TensorShape([2, 3, 2])
"""
mask = tf.Variable([
[1, 1, 0],
[1, 0, 0]
])
# mask dtype must be boolean
mask = tf.cast(mask, dtype=bool)
# `mask`'s shape must be a prefix of `data`'s shape
mask.shape
"""
TensorShape([2, 3])
"""
# if we don't mask
tf.reduce_mean(var, axis=1)
"""
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[0.76666665, 0.76666665],
[0.53333336, 0.53333336]], dtype=float32)>
"""
# what the ragged tensor looks like:
tf.ragged.boolean_mask(var, mask=mask)
"""
<tf.RaggedTensor
[
[[0.5, 0.5], [0.8, 0.8]],
[[0.2, 0.2]]
]>
"""
# result after mask:
tf.reduce_mean(tf.ragged.boolean_mask(var, mask=mask), axis=1)
"""
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[0.65, 0.65],
[0.2 , 0.2 ]], dtype=float32)>
"""
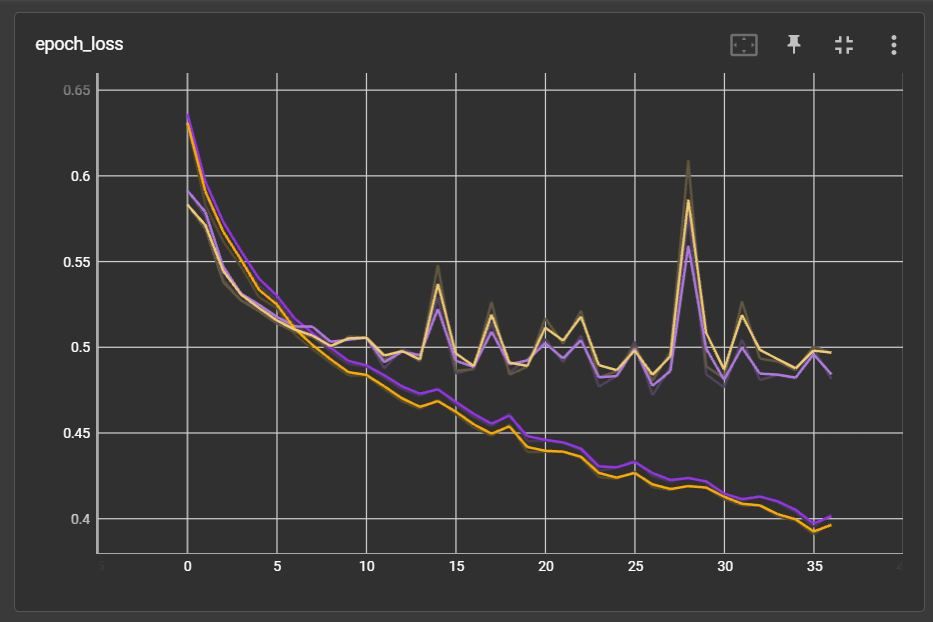
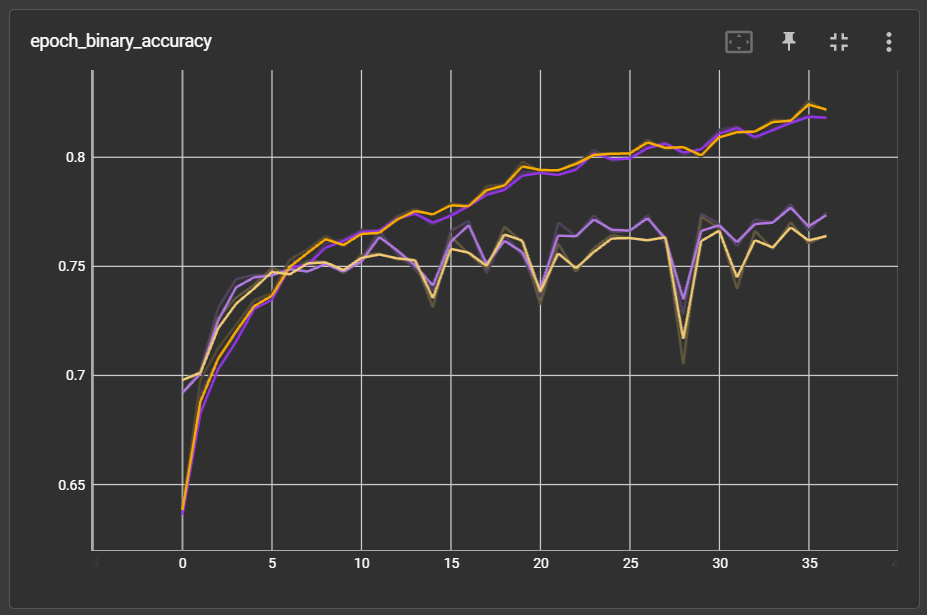
The performance did improve slightly. With same data split and weight initialization, model on masked embedding (purple) has slightly lower validation loss and higher validation accuracy: